Bring Vector Compression to the Extreme: How Milvus Serves 3× More Queries with RaBitQ
Milvus is an open-source, highly scalable vector database powering semantic search at a billion-vector scale. As users deploy RAG chatbots, AI customer service, and visual search at this magnitude, a common challenge emerges: infrastructure costs. In contrast, exponential business growth is exciting; skyrocketing cloud bills are not. Fast vector search typically requires storing vectors in memory, which is expensive. Naturally, you might ask: Can we compress vectors to save space without sacrificing search quality?
The answer is YES, and in this blog, we’ll show you how implementing a novel technique called RaBitQ enables Milvus to serve 3× more traffic with lower memory cost while maintaining comparable accuracy. We’ll also share the practical lessons learned from integrating RaBitQ into open-source Milvus and the fully-managed Milvus service on Zilliz Cloud.

Understanding Vector Search and Compression
Before diving into RaBitQ, let’s understand the challenge.
Approximate Nearest Neighbor (ANN) search algorithms are at the heart of a vector database, finding the top-k vectors closest to a given query. A vector is a coordinate in high-dimensional space, often comprising hundreds of floating-point numbers. As vector data scales up, so do storage and compute demands. For instance, running HNSW (an ANN search algorithm) with one billion 768-dimensional vectors in FP32 requires over 3TB of memory!
Like MP3 compresses audio by discarding frequencies imperceptible to the human ear, vector data can be compressed with minimal impact on search accuracy. Research shows that full-precision FP32 is often unnecessary for ANN. Scalar Quantization (SQ), a popular compression technique, maps floating-point values into discrete bins and stores only the bin indices using low-bit integers. Quantization methods significantly reduce memory usage by representing the same information with fewer bits. Research in this domain strives to achieve the most savings with the least loss in accuracy.
The most extreme compression technique—1-bit Scalar Quantization, also known as Binary Quantization—represents each float with a single bit. Compared to FP32 (32-bit encoding), this reduces memory usage by 32×. Since memory is often the main bottleneck in vector search, such compression can significantly boost performance. The challenge, however, lies in preserving search accuracy. Typically, 1-bit SQ reduces recall to below 70%, making it practically unusable.
This is where RaBitQ stands out—an excellent compression technique that achieves 1-bit quantization while preserving high recall. Milvus now supports RaBitQ starting from version 2.6, enabling the vector database to serve 3× the QPS while maintaining a comparable level of accuracy.
A Brief Intro to RaBitQ
RaBitQ is a smartly designed binary quantization method that leverages the geometry property of high-dimensional space to achieve efficient and accurate vector compression.
At first glance, reducing each dimension of a vector to a single bit may seem too aggressive, but in high-dimensional space, our intuitions often fail us. As Jianyang Gao, an author of RaBitQ, illustrated, high-dimensional vectors exhibit the property that individual coordinates tend to be tightly concentrated around zero, a result of a counterintuitive phenomenon explained in Concentration of Measure. This makes it possible to discard much of the original precision while still preserving the relative structure needed for an accurate nearest neighbor search.
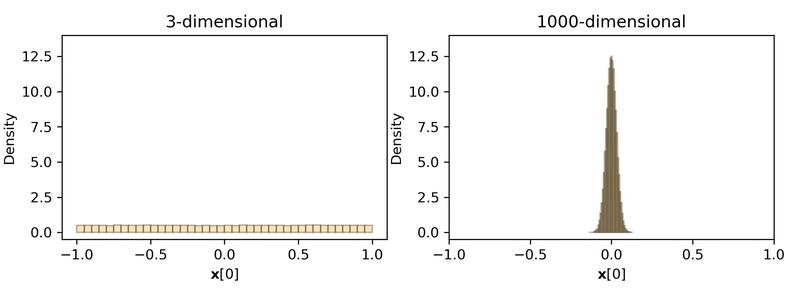
Figure: The counterintuitive value distribution in high-dimensional geometry. Consider the value of the first dimension for a random unit vector uniformly sampled from the unit sphere; the values are uniformly spread in 3D space. However, for high-dimensional space (e.g., 1000D), the values concentrate around zero, an unintuitive property of high-dimensional geometry. (Image source: Quantization in The Counterintuitive High-Dimensional Space)
Inspired by this property of high-dimensional space, RaBitQ focuses on encoding angular information rather than exact spatial coordinates. It does this by normalizing each data vector relative to a reference point such as the centroid of the dataset. Each vector is then mapped to its nearest vertex on the hypercube, allowing representation with just 1 bit per dimension. This approach naturally extends to IVF_RABITQ, where normalization is done relative to the closest cluster centroid, improving local encoding accuracy.
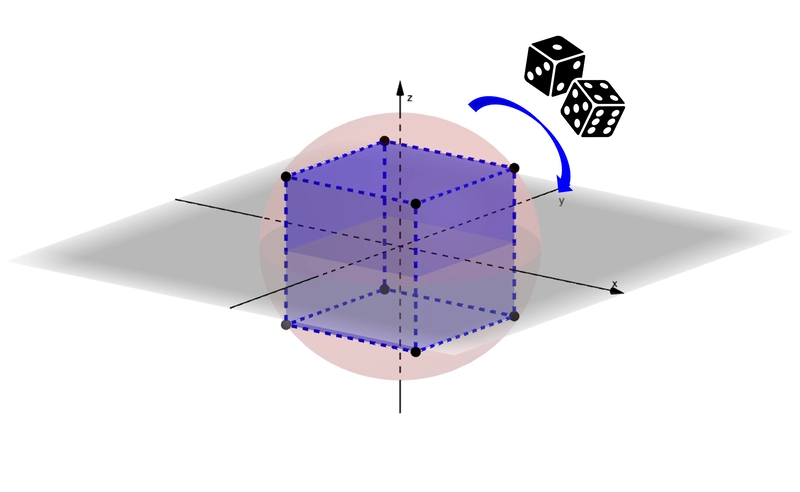
Figure: Compressing a vector by finding its closest approximation on the hypercube, so that each dimension can be represented with just 1 bit. (Image source: Quantization in The Counterintuitive High-Dimensional Space)
To ensure search remains reliable even with such compressed representations, RaBitQ introduces a theoretically grounded, unbiased estimator for the distance between a query vector and binary-quantized document vectors. This helps minimize reconstruction error and sustain high recall.
RaBitQ is also highly compatible with other optimization techniques, such as FastScan and random rotation preprocessing. Moreover, RaBitQ is lightweight to train and fast to execute. Training involves simply determining the sign of each vector component, and search is accelerated via fast bitwise operations supported by modern CPUs. Together, these optimizations enable RaBitQ to deliver high-speed search with minimal loss of accuracy.
Engineering RaBitQ in Milvus: From Academic Research to Production
While RaBitQ is conceptually straightforward and accompanied by a reference implementation, adapting it in a distributed, production-grade vector database like Milvus presented several engineering challenges. We’ve implemented RaBitQ in Knowhere, the core vector search engine behind Milvus, and also contributed an optimized version to the open-source ANN search library FAISS.
Let’s look at how we brought this algorithm to life in Milvus.
Implementation Tradeoffs
One important design decision involved handling per-vector auxiliary data. RaBitQ requires two floating-point values per vector pre-computed during indexing time, and a third value that can either be computed on-the-fly or pre-computed. In Knowhere, we pre-computed this value at indexing time and stored it to improve efficiency during search. In contrast, the FAISS implementation conserves memory by calculating it at query time, taking a different tradeoff between memory usage and query speed.
Hardware Acceleration
Modern CPUs offer specialized instructions that can significantly accelerate binary operations. We tailored the distance computation kernel to take advantage of modern CPU instructions. Since RaBitQ relies on popcount operations, we created a specialized path in Knowhere that uses the VPOPCNTDQ instructions for AVX512 when available. On supported hardware (e.g., Intel IceLake or AMD Zen 4), this can accelerate binary distance computations by several factors compared to default implementations.
Query Optimization
Both Knowhere (Milvus’s search engine) and our optimized FAISS version support scalar quantization (SQ1–SQ8) on query vectors. This provides additional flexibility: even with 4-bit query quantization, recall remains high while computational demands decrease significantly, which is particularly useful when queries must be processed at high throughput.
We go a step further in optimizing our proprietary Cardinal engine, which powers the fully managed Milvus on Zilliz Cloud. Beyond the capabilities of the open-source Milvus, we introduce advanced enhancements, including integration with a graph-based vector index, additional layers of optimization, and support for Arm SVE instructions.
The Performance Gain: 3× More QPS with Comparable Accuracy
Starting with version 2.6, Milvus introduces the new IVF_RABITQ index type. This new index combines RaBitQ with IVF clustering, random rotation transformation, and optional refinement to deliver an optimal balance of performance, memory efficiency, and accuracy.
Using IVF_RABITQ in Your Application
Here’s how to implement IVF_RABITQ in your Milvus application:
from pymilvus import MilvusClient
index_params = MilvusClient.prepare_index_params()
index_params.add_index(
field_name="your_vector_field_name", # Name of the vector field to be indexed
index_type="IVF_RABITQ", # Will be introduced in Milvus 2.6
index_name="vector_index", # Name of the index to create
metric_type="IP", # IVF_RABITQ supports IP and COSINE
params={
"nlist": 1024, # IVF param, specifies the number of clusters
} # Index building params
)
Benchmarking: Numbers Tell the Story
We benchmarked different configurations using vdb-bench, an open-source benchmarking tool for evaluating vector databases. Both the test and control environments use Milvus Standalone deployed on AWS EC2 m6id.2xlarge instances. These machines feature 8 vCPUs, 32 GB of RAM, and an Intel Xeon 8375C CPU based on the Ice Lake architecture, which supports the VPOPCNTDQ AVX-512 instruction set.
We used the Search Performance Test from vdb-bench, with a dataset of 1 million vectors, each with 768 dimensions. Since the default segment size in Milvus is 1 GB, and the raw dataset (768 dimensions × 1M vectors × 4 bytes per float) totals roughly 3 GB, the benchmarking involved multiple segments per database.
Figure: Example test configuration in vdb-bench.
Here are some low level details about the configuration knobs for IVF, RaBitQ and refinement process:
nlistandnprobeare standard parameters for allIVF-based methodsnlistis a non-negative integer that specifies the total number of IVF buckets for the dataset.nprobeis a non-negative integer that specifies the number of IVF buckets that are visited for a single data vector during the search process. It is a search-related parameter.rbq_bits_queryspecifies the level of quantization of a query vector. Use 1…8 values for theSQ1…SQ8levels of quantization. Use 0 value to disable quantization. It is a search-related parameter.refine,refine_typeandrefine_kparameters are standard parameters for the refine processrefineis a boolean that enables the refinement strategy.refine_kis a non-negative fp-value. The refining process uses a higher quality quantization method to pick the needed number of nearest neighbors from arefine_ktimes larger pool of candidates, chosen usingIVFRaBitQ. It is a search-related parameter.refine_typeis a string that specifies the quantization type for a refining index. Available options areSQ6,SQ8,FP16,BF16andFP32/FLAT.
The results reveal important insights:
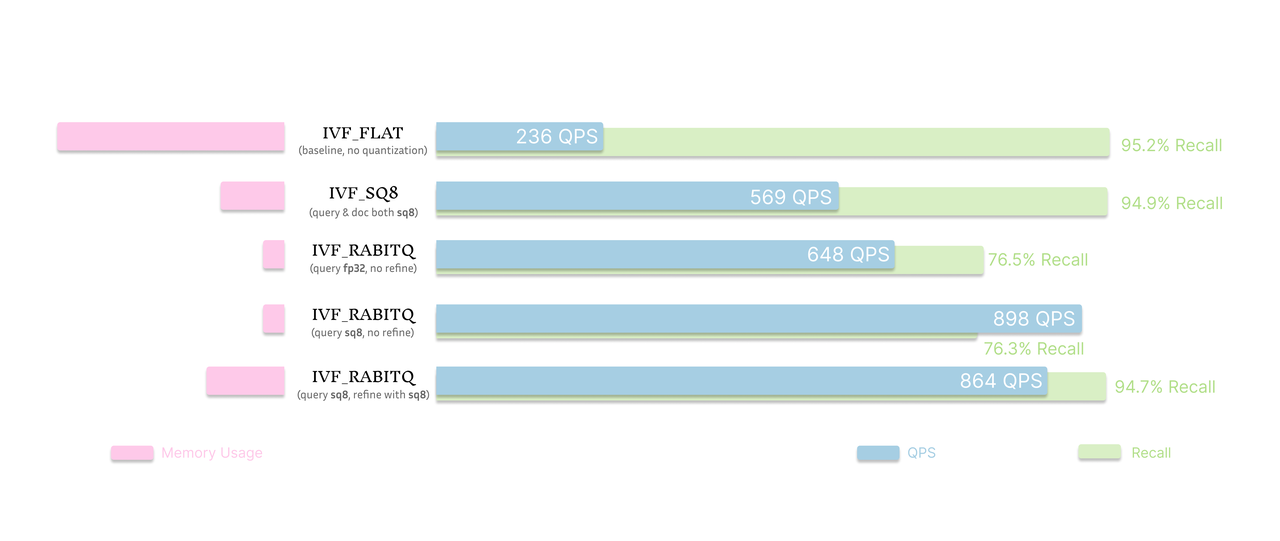
Figure: Cost and performance comparison of baseline (IVF_FLAT), IVF_SQ8 and IVF_RABITQ with different refinement strategies
Compared to the baseline IVF_FLAT index, which achieves 236 QPS with 95.2% recall, IVF_RABITQ reaches significantly higher throughput—648 QPS with FP32 queries and 898 QPS when paired with SQ8-quantized queries. These numbers demonstrate the performance advantage of RaBitQ, especially when refinement is applied.
However, this performance comes with a noticeable trade-off in recall. When IVF_RABITQ is used without refinement, recall levels off at around 76%, which may fall short for applications that require high accuracy. That said, achieving this level of recall using 1-bit vector compression is still impressive.
Refinement is essential for recovering accuracy. When configured with SQ8 query and SQ8 refinement, IVF_RABITQ delivers both great performance and recall. It maintains a high recall of 94.7%, nearly matching IVF_FLAT, while achieving 864 QPS, over 3× higher than IVF_FLAT. Even compared with another popular quantization index IVF_SQ8, IVF_RABITQ with SQ8 refinement achieves more than half of the throughput at similar recall, only with a marginal more cost. This makes it an excellent option for scenarios that demand both speed and accuracy.
In short, IVF_RABITQ alone is great for maximizing throughput with acceptable recall, and becomes even more powerful when paired with refinement to close the quality gap, using only a fraction of the memory space compared to IVF_FLAT.
Conclusion
RaBitQ marks a significant advancement in vector quantization technology. Combining binary quantization with smart encoding strategies, it achieves what seemed impossible: extreme compression with minimal accuracy loss.
Starting with version 2.6, Milvus will introduce IVF_RABITQ, integrating this powerful compression technique with IVF clustering and refinement strategies to bring binary quantization to production. This combination creates a practical balance between accuracy, speed, and memory efficiency that can transform your vector search workloads.
We’re committed to bringing more innovations like this to both open-source Milvus and its fully managed service on Zilliz Cloud, making vector search more efficient and accessible for everyone.
Getting Started with Milvus 2.6
Milvus 2.6 is available now. In addition to RabitQ, it introduces dozens of new features and performance optimizations such as tiered storage, Meanhash LSH, and enhanced full-text search and multitenancy, directly addressing the most pressing challenges in vector search today: scaling efficiently while keeping costs under control.
Ready to explore everything Milvus 2.6 offers? Dive into our release notes, browse the complete documentation, or check out our feature blogs.
If you have any questions or have a similar use case, feel free to reach out to us through our Discord community or file an issue on GitHub — we’re here to help you make the most of Milvus 2.6.
- Understanding Vector Search and Compression
- A Brief Intro to RaBitQ
- Engineering RaBitQ in Milvus: From Academic Research to Production
- Implementation Tradeoffs
- Hardware Acceleration
- Query Optimization
- The Performance Gain: 3× More QPS with Comparable Accuracy
- Using IVF_RABITQ in Your Application
- Benchmarking: Numbers Tell the Story
- Conclusion
- Getting Started with Milvus 2.6
On This Page
Try Managed Milvus for Free
Zilliz Cloud is hassle-free, powered by Milvus and 10x faster.
Get StartedLike the article? Spread the word